There was a time when it seemed that medicine and human ingenuity could remove deadly infections from the face of the planet. The stunning success of vaccines in wiping out smallpox and polio and of antibiotic drugs in fighting infections suggested that, with the right molecules and strategies, humans could rid themselves of any scourge. But that view, increasingly, has been altered by a reality of evolutionary biology: everything changes and adapts, and microbes have a way of outmaneuvering medicine.
The evolution of microbes has resulted in SARS, Ebola hemorrhagic fever, AIDS, and swine flu: diseases that have jumped to humans from animal hosts. In the case of older diseases, resistance to antimicrobial drugs is now a common occurrence. Increased global travel means that new infections and “bugs” possessing drug resistance can leap to another part of the world overnight.
One of the clear cases is malaria. Several decades ago, the world was poised to celebrate the eradication of this disease, but ever since, cases have been on the rise. Every year, more than a million people—mostly children in Africa—die of malaria, while hundreds of millions are infected but survive. Only a handful of antimalarial drugs exist, and no new drugs have been made available in decades. Not only is microbial resistance to the cheapest and most widely used drug, chloroquine, already widespread in endemic areas, but resistance to a combination treatment, sulfadoxine-pyrimethamine, which first appeared in the 1980s in South America and Southeast Asia, has now spread to East Africa. Resistance to artemisinin, the newest antimalarial (first developed in the 1970s), could also be emerging.
Fortunately, understanding how pathogens evolve can shed new light on the dynamics of disease. Dyann F. Wirth, Strong professor of infectious disease, who chairs the department of immunology and infectious diseases at the Harvard School of Public Health (HSPH), has been studying how and why drug resistance emerges in malaria. “Drug resistance is just a special case of evolutionary biology,” she says. Wirth, who directs the Harvard Malaria Initiative (see sidebar), is leading an effort to use new genomic tools, first developed for studying evolution, as a means to better understand malaria and find effective ways to fight it.
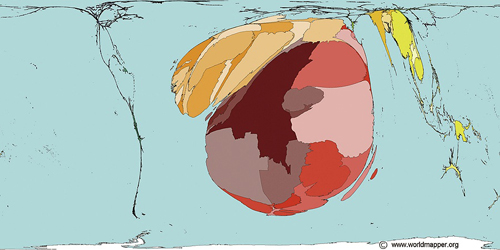
Map Copyright © 2006 SASI Group (University of Sheffield) and Mark Newman (University of Michigan)
Deaths from malaria worldwide, as shown on a map resized to reflect the proportional burden of mortality. The majority of deaths and cases are concentrated in Africa and Southeast Asia.
Reconstructing evolution
Malaria has an impressive ability to evolve in response to challenges. In fact, the malaria protozoan’s ability to adapt rapidly mimics the behavior of viruses more than that of other single-celled organisms. Treating the disease with a drug immediately prompts its evolution toward developing resistance. The world has seen this phenomenon in action with chloroquine, a synthetic drug that replaced natural quinine as the antimalarial of choice after World War II. Chloroquine resistance emerged separately in Southeast Asia and South America more than 50 years ago, and scientists can trace its origin to mutations in a specific gene. Gradually, the resistant types came to dominate. “You can trace the history of the spread of chloroquine resistance with the spread of this resistant locus within the genome,” Wirth says.
Though she has been studying malaria throughout her career, Wirth began thinking of her work more formally as evolutionary biology a decade ago, when she began collaborating with Higgins professor of biology Daniel L. Hartl. An evolutionary biologist himself, Hartl was interested in knowing how much genetic variation existed in the protozoan that causes malaria, because, he says, the parasite is “a very strange creature with a hugely complex life cycle, and not a whole lot was known about it 10 years ago when we started this.”
There are four species of single-celled protozoans that cause malaria in humans. The most widespread, Plasmodium vivax, is found in Asia, North Africa, the Middle East, and South and Central America; the most deadly, Plasmodium falciparum, is concentrated in southern Africa. The parasite travels back and forth between its two hosts, humans and mosquitoes, and malaria is spread through mosquito bites. Malaria can mean many things: an asymptomatic infection, a brief fever with chills and aches, or severe anemia or disease in the kidneys and brain, which can rapidly lead to death. The victims are usually children; most individuals who reach adolescence are able to coexist in relative peace with the parasite thereafter.
Malaria parasites enter the human body from the salivary glands of a feeding mosquito. They move to the liver, replicate rapidly, and then invade the red blood cells, where they replicate further. Most continue to multiply in this way, infecting more blood cells, but a small subset develops into male and female gametocytes. When next a mosquito feasts on the human host’s blood, some of these gametocytes may be drawn into the mosquito’s gut, where they reproduce sexually—a step that distinguishes malaria from bacterial and viral pathogens. The resulting offspring then divide and move to the salivary glands, where the process begins anew. This unusual cycling back and forth is one of the keys to malaria’s rapid evolution. “Every time the parasite is transmitted, you essentially have a mating event,” Wirth says. “And if you think about Africa, where people have been bitten every night by multiple mosquitoes, you can imagine that there’s an enormous amount of exchanging genetic information.” All this suggested to her that there would be a great deal of genetic diversity among parasites and a great deal of variation in the malaria genome.
When the Broad Institute of MIT and Harvard was launched in 2004, its director, Eric Lander, professor of systems biology at Harvard Medical School (HMS), asked Wirth to co-lead its infectious disease program with HMS professor of biological chemistry and molecular pharmacology Jon Clardy. Wirth decided to take a yearlong sabbatical at the Broad to better understand large-scale genomics and to find a way to apply the tools originally developed for the Human Genome Project to malaria. Because the P. falciparum genome is only one three-hundredth the size of the human genome, she points out, applying these tools yields powerful results much more quickly.
The genome sequence of Plasmodium falciparum, consisting of the entire sequence of DNA molecules in a single parasite, had been completed in 2002, but a single genome cannot indicate how the parasite has evolved over time and how diverse the parasites are across different geographical regions. One of the group’s first tasks was to get a picture of the genetic diversity of P. falciparum by gathering genomic information from many different parasites. The human Haplotype Map (HapMap) project was launched in 2002 to create a comprehensive map of common genetic variation in humans; Wirth’s team at the Broad led a similar effort with colleagues in Dakar, Senegal, to create a genome-wide map of genetic diversity in P. falciparum. By partially sequencing the genomes of 16 parasites from different parts of the world, they were able to create a map of the most common genetic variations in the species—information they could use to further analyze more than 50 other parasites to address questions about parasite diversity and evolution. The map showed that the malaria genome was even more diverse than Wirth expected.
Pardis Sabeti, a former postdoctoral fellow at the Broad who is now assistant professor in the department of organismic and evolutionary biology (see “Harvard Portrait,” May-June 2009, page 49), and Sarah K. Volkman, a senior research scientist in Wirth’s lab, then helped lead an effort to understand how malaria has evolved. To do this, the scientists took advantage of a measure called linkage disequilibrium, which determines how often one part of the genome is linked to another. Genomes get shuffled over time; every time parasites mate, some of their genes can switch to another chromosome, a process called recombination. Just as shuffling a deck of cards reduces the likelihood of seeing the same card sequences, recombination makes it less likely that any one variation will always be seen in sequence with any other. Linkage disequilibrium thus tells scientists how frequently different sites on the genome are linked to one another, and gives them a measure of evolutionary age: traits that are more often linked have been shuffled less and so have emerged more recently.
This measure also tells scientists something about the selective pressures that are acting on the organism. Genes that have emerged more recently reflect traits that are under selection by their environment—traits that allow the organism to adapt and survive. “This allows us to begin to look for signatures of selection,” Wirth says. “We’re essentially sequencing the parasites that exist today and inferring their population history.” This approach can also be used in reverse, to understand which genes are responsible for a recent evolutionary change. In the case of drug resistance, Wirth says, scientists look for less diverse areas of the genome to track recently emerged genes that may have made a given parasite resistant.
It’s possible to trace the history not only of an entire genome, but also of a single gene. Daniel Hartl, for example, became interested in reconstructing the evolutionary history of individual cases of microbial resistance to malarial drugs. His lab published a paper last summer that looked closely at the emergence of resistance to pyrimethamine, which involved mutations in four amino acids that form the building blocks of a particular enzyme in the parasite P. falciparum. The researchers were able to reconstruct the order in which the amino acids changed by recreating possible scenarios using bacteria engineered with different forms of the enzyme. “I was interested in this from two points of view,” Hartl says. One was practical: if scientists know how resistance evolved, they can begin to look for early stages of resistance in other parasites. The other, he explains, “is a much more general evolutionary question, which is, ‘How do proteins evolve?’” Many evolutionary adaptations require multiple mutations to take place, and it’s been unclear until recently how these intermediate steps occur.
A battle between host and parasite
The availability of genomic technologies has allowed Wirth’s team not only to look in detail at drug resistance, but also to get an overall picture of the forces that have shaped the malaria genome. And it turns out that the most important influence is the human body.
“I was expecting drug resistance genes to pop out as the genes under the highest selection by the parasite’s environment,” Wirth says. Instead, they found that the parasite demonstrates incredible diversity in the genes that encode proteins on the protozoan’s surface—the molecules that are typically recognized by the human immune system. “What this told us immediately,” she explains, “was that these genes were likely to be targets of the human immune response, and that variation to escape the immune system was probably the important force in driving the evolution of parasite populations in the world.” In hindsight, Wirth says, she should have predicted as much, because antimalarial drugs have been widely used for less than 100 years, and many people infected with malaria are never treated at all. In contrast, “The human immune system is subjecting the parasite to selection 24/7,” and has been doing so for thousands of years, so “you almost have an arms race between the parasite and the host.”
In this race, however, the parasite has many more opportunities to adapt. Wirth points out that at every step of its life cycle, “the parasite is always facing elimination.” Though people may carry billions of malaria protozoans in their blood, a biting mosquito picks up only a few that are capable of reproducing. These few multiply in the mosquito’s body, and when that mosquito bites again, only a few of the hundreds of thousands of parasites that it carries are injected into its human victim. As Wirth says, “It would be like taking the Harvard medical area and saying that only one person is going to survive some hypothetical selection process. If you think about it from a competitive standpoint, that’s going to be the person best adapted to whatever the test is.”
In Africa, P. falciparum has textbook conditions for an evolutionary perfect storm. With its two hosts living in close proximity, the parasite has many opportunities to mate and diversify, but the bottlenecks it faces as it moves between mosquito and human ensure that only the best adapted individuals survive. This abundant variation, when combined with stringent selection, fosters rapid evolution.
But humans have adapted too, in our own relatively sluggish way. Although malaria has been with us for hundreds of thousands of years, the parasite’s numbers exploded around the time agriculture was developed 10,000 years ago. Humans were then living in denser populations and clearing land for fields and settlements, which meant that some of them were living close to wet habitats where mosquitoes bred. Much of the malaria genome evolved after this large expansion, which was also when the mosquito species that transmit malaria began to expand and diversify.
Scientists looking at the human genome can see that malaria has been one of the most important factors in shaping it ever since. The disease has a direct effect on human survival and reproduction because infection causes complications in pregnancy and also because malaria’s victims are usually children. In fact, it was human adaptation to malaria that provided the first direct evidence that the human race has indeed evolved. In 1949—more than 75 years after Darwin argued in The Descent of Man that humans were products of natural selection—sickle cell anemia was found to arise from a genetic adaptation that protects people from malaria. Other blood disorders have subsequently been linked to malaria resistance as well, which helps explain why these seemingly harmful genetic diseases have become prevalent among people living in tropical areas.
Pardis Sabeti has looked at both host and parasite genomes to trace the battle scars of this relationship. Working with Eric Lander, she first developed a method for identifying artifacts of natural selection in the human genome in 2002. In subsequent years she has been able to confirm several known recent adaptations in humans, including the gene that causes sickle cell anemia. She has also helped look for evidence of natural selection in malaria. Sabeti believes that understanding both genomes will help scientists understand the clinical course of the disease better. “If someone becomes ill from malaria,” she points out, “it could either be that the person does not have good immunity to the parasite or it could be that the infecting parasite is particularly severe. So you really have to look at both sides.”
Of course, there are actually three players in this evolutionary dance. Mosquitoes have also evolved resistance to insecticides, so understanding their evolution is important as well. The mosquito genome has also been sequenced, and the Harvard team plans to apply some of the same genomic tools to better understand its role in malaria. In particular, the insect’s genome could provide insight into differences in malaria between Southeast Asia and Africa, where different mosquito species transmit the disease.
From laboratory to public health
The ability to trace the evolution of malaria is not a mere academic exercise. The genomic information generated by this work can be used to identify and track drug resistance in countries where malaria is endemic. The malaria HapMap project made it possible to take some of the key variations found in Plasmodium genomes and use them as a way to identify the parasites in infected patients.
A team based at the Broad Institute developed a set of 24 genetic markers that could be used to create a molecular “bar code” that could uniquely identify any parasite in a patient’s blood. The markers are embedded in a chip that can be manufactured inexpensively and used with standard lab equipment. Wirth says the technology can be used in the clinic to better characterize and track a particular patient’s disease. Although it’s possible to track resistance to a single drug with other methods, the molecular bar code would make it possible to test for resistance to multiple drugs at once and identify which drug will work for a specific parasite.
Wirth and her colleagues have worked with Senegalese scientists from the University of Cheikh Anta Diop, in Dakar, to study both the genetic diversity of malaria in that country and microbial drug resistance among malaria parasites in patients. Cases of chloroquine resistance were first reported in the 1980s in Senegal and are now common there. The team has therefore been studying the underlying genetic mutations responsible for making the parasite resistant to treatment, with the goal of eventually using a molecular bar-code system to identify and track emerging cases of resistance to newer drugs. Wirth says that they have already seen resistance to pyrimethamine appear in Senegal and spread gradually as the resistant protozoan took over the parasite population. They hope to focus now on artemisinin, the newest antimalarial, which is used in combination with others. “The goal would be to catch artemisinin resistance much earlier, so we could perhaps contain it,” she explains. Early warning of resistance would make it possible to direct resources like insecticides where they are needed most. Wirth notes that—apart from the consequences a drug-resistant infection has for an individual patient—“the issue from a public-health standpoint is that when a resistant organism begins to spread, you have to completely change the public-health approach.”
The collaboration in Senegal has also provided a setting for investigating new questions about malaria. Johanna Daily, a former member of Wirth’s lab who is now an associate professor of medicine at Albert Einstein College, led a 2007 study that used a genomic tool to identify which genes are active in parasites taken directly from patients’ blood, as distinct from parasites cultured in a lab. That research showed that some of the blood parasites were operating in two totally different biological states: one apparently a response to an environmental stress; the other a response to the denial of typical food sources. The study points to a way to correlate the parasite’s physical state with a patient’s symptoms—which could clarify why the severity of infection varies so widely. Wirth says that Senegal will provide an opportunity for the team to look in much more depth at parasite diversity and to explore its relationship to human disease and mosquito biology.
The diversity of the parasite may play a role not just in drug resistance but in the nature of the disease itself. Dan Milner, a pathologist at Brigham and Women’s Hospital, is working with Wirth, Michigan State University researcher Terrie Taylor, and colleagues at the Queen Elizabeth Central Hospital in Blantyre, Malawi, to investigate cases of fatal malaria infection to understand what causes the most severe forms of the disease. In seeking to characterize the diversity of malaria parasites in Malawi, he hopes to learn whether genetic differences among them contribute to disease severity. In Malawi, he says, many more people die of malaria than in Senegal.
Severe malaria is usually seen in young children. Gradually, most people develop a tolerance for the disease; they may still be infected many times by malaria protozoans, but their bodies are able to control the infection. Milner says this tolerance depends on a continual back-and-forth between the parasites and the human immune system. In areas where the disease is endemic, he says, infection is “like a natural constant vaccination.” But those who leave Malawi to spend a few years abroad no longer have this immunity when they return, because the parasites have changed so much in the interim.
Vaccines, new drugs, bed nets, and more
Wirth believes that malaria’s remarkable diversity has implications for the development of vaccines. So far there is no effective vaccine for malaria; Wirth says that many candidates work well in animal studies but fail when tested in humans. She notes that many vaccines developed so far target the very features of the parasite that are most variable, and therefore they may not work on all parasites. Instead, she suggests, it may be necessary to develop pan-vaccines—like those used for influenza—that are modified every year to attack the viruses predicted to be most common. Although this annual forecasting approach might work in theory, its expense might prove too great for the practical realities of poor countries where malaria is endemic. Wirth’s group, therefore, is pursuing a different strategy: focusing on identifying possible vaccine targets that represent critical, less variable functions in the parasite.
With or without a vaccine, controlling malaria will be a significant challenge. In the 1950s and 1960s, widespread campaigns to eradicate the disease were conducted using newly developed drugs (against the malarial parasite) and DDT (against the mosquitoes that host it). Countries around the world celebrated the eradication of the disease. But shortly afterward, resistance emerged in both the parasite and the mosquitoes and the disease surged again. Although eradicated from the United States and other developed countries, malaria kills more people today than ever; recently, the Bill and Melinda Gates Foundation has called for a renewed effort to eradicate the parasite globally.
Current efforts against malaria focus on a multipronged strategy. A malaria vaccine, if one is developed, could help prevent or at least slow infections. For those who do become infected, the development of new treatments would ensure that recovery remains possible even as older drugs become less effective. Mosquito control, when deployed strategically, can also prevent infections at their source. And the low-tech approach of using bed nets in endemic areas also has a great potential for stopping malaria before it starts. “I don’t think any one of these things will do very much,” Dan Hartl says, “but all together they should have a very significant effect.”
Even if controlling the disease is a realistic goal, Hartl believes it may be naïve to think we can wipe malaria from the face of the earth, given its incredible ability to adapt. Awareness is growing about the evolutionary reality of infectious diseases like malaria, he says: “It’s important to take the long view, because in my opinion, malaria is going to be with the human population for a very, very long time.” And malaria is just one example of this new reality—treating HIV also requires varied combinations of antiretroviral drugs. Fighting a disease is no longer a matter of a single vaccine or drug; it must include an ever-changing arsenal of weapons: a steady stream of new antibiotics to combat resistant bacteria, vaccines that adapt to changing pathogens, and a more complete knowledge of the enemy’s most fundamental weaknesses.









