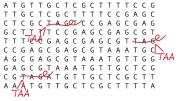Scientists have made stunning progress in their ability to decode genomes; the past several years have produced many new genome-sequencing efforts. Now, some scientists have shifted their focus from passively reading genomes to actively “writing” and “editing” them in specific ways. Researchers led by George Church, professor of genetics at Harvard Medical School, and Joe Jacobson, associate professor in the Media Lab at MIT, have announced a new approach for rapidly and inexpensively editing large numbers of genomes in living cells. Their new editing tools could be used to engineer cells that have radically different properties, including advantages such as resistance to infection.
Their July 15 paper in Science focuses on efforts to alter the genome by means of a “search and replace” method that revises codons—strings of three DNA molecules that are often thought of as DNA “words” because they encode a single amino acid (the building block of proteins). Some codons, though, function more like a punctuation mark; these “stop codons” instruct the protein-building machinery of a cell to stop linking amino acids together, much as a period ends a sentence. In this study, the researchers engineered E. coli bacteria by replacing each stop codon bearing the pattern of DNA bases thymine-adenine-guanine, or TAG, with a different (but nonharmful) stop codon reading TAA.
Replacing each instance of that single codon, which appears 314 times in the E. coli genome, involved a two-step process. “We’re speeding up evolution,” Church explains. The first technique, called multiplex automated genome engineering (MAGE), can make rapid, specific changes in cells: researchers introduce pieces of synthetic DNA into bacteria and then select resulting strains that possess the desired properties (in this case, with some of the TAG codons replaced by TAA). They then use a method called conjugative assembly genome engineering (CAGE) that draws on bacteria’s natural ability to swap genetic material. By selecting strains with the most TAA codons, the team could eventually create a strain in which every instance of the TAG codon has been replaced.
The larger goal, Church says, is to create a bacterium that is resistant to infection by viruses. Viruses can’t make their own proteins, so they hijack host genomes and force them to make viral proteins; Church’s team wants to scramble the genetic code in a way that leaves its functions intact but makes it unrecognizable to viral invaders. The TAG and TAA codons are “synonymous”—they both have the same function in the genome—so by replacing each TAG stop codon with a TAA equivalent, Church’s team can then delete the machinery in the cell responsible for reading TAG. The cell will function as if everything is normal, but a virus expects to see the code in its original form. “If you make the code different enough,” Church explains, “that organism becomes resistant to all viruses.”
Viral resistance is only one of the ways these editing techniques can be used to hack genomes. “Our goal is to change every base pair,” Church reports. His approach differs from that of a team at the J. Craig Venter Institute that synthesized a genome from scratch and transplanted it into a cell last year. Rather than copy something from life, Church says, his team’s goal is to radically change an existing genome’s properties. He estimates that creating the entire E. coli genome would cost several million dollars, whereas his editing technique is much cheaper and more flexible, and at each stage of the process provides researchers with feedback about how their changes have affected the cell. With a directed evolution approach, he says, “If you select for what you want, the end product is exactly what you want.”