For thousands of ordinary people around the world, one of biology’s hardest problems is just a game. Both scientists and supercomputers have long struggled to predict the three-dimensional structures of the biological molecules called proteins. These structures are crucial to understanding proteins’ roles in fundamental cellular processes and disease, but predicting them is no easy task—which is why some researchers have turned to laypeople for help.
In theory, a protein’s structure should be calculable from the molecule’s underlying chemistry: from its initial state as a linear chain of chemical building blocks called amino acids, each protein is thought to fold into its most stable possible configuration. But there are infinite structural possibilities for any given amino-acid chain, and a computer, searching through them, faces a daunting challenge.
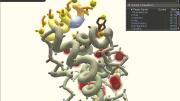
In the early 2000s, David Baker ’84, a biochemistry professor at the University of Washington (UW), Seattle, launched a project called Rosetta@home to outsource the critical scientific work of protein structure prediction from supercomputers to thousands of idle home computers. An algorithm, Rosetta, sifted through the many possibilities while a screensaver showing the various protein-folding permutations kept users updated on its progress.
Then something unexpected happened. Before long, “People started writing in, saying, ‘I can see where it would fit better this way,’” Baker told the journal Nature in 2010. With that, the Baker lab and researchers from UW’s computer-science department began exploring a second possibility: making it possible for those frustrated Rosetta@home hosts to fold proteins on their own. The scientists designed an interface that let users move amino acids with the click of a mouse, and they embedded tools with names like “wiggle” and “shake” that could adjust entire regions of a protein at once. The result was Foldit, a game that let nonprofessionals try their hands at protein-folding problems that had stymied supercomputers.
In 2008, the developers released the game and invited ordinary citizens to play.
Foldit is part of a growing trend toward citizen science: enabling ordinary people, often without formal training, to contribute to scientific research in their spare time. The range of involvement varies. Some citizen scientists donate idle time on their home computers for use in solving problems large in scale (the search for intergalactic objects, as in Einstein@home) or small (folding proteins). Other projects encourage participants to contribute small bits of data about themselves or their environments. The Great Sunflower Project, for instance, provides a platform for logging and sharing observations of pollinators like bees and wasps. Still other efforts enlist laypeople to tag and analyze images: Eyewire, for example, a game developed by Sebastian Seung ’86, Ph.D. ’90, a professor of computational neuroscience at MIT, involves participants in mapping neurons in the brain.
“There’s a good, long history of people in orthodox scientific domains enrolling members of the public,” says Sheila Jasanoff, Pforzheimer professor of science and technology studies at Harvard Kennedy School. In the eighteenth and nineteenth centuries, amateur naturalists like England’s Gilbert White played an important role in cataloging local flora and fauna. Active lay communities still exist in fields like astronomy and ornithology, she notes, and frequently, citizen science simply organizes what people already do.
But the Internet and mobile phones now connect more people than ever before, changing how scientists and citizens interact. Today’s citizen science is born from and reinforces other shifts in the digital world—“big data,” open access, and mobile-phone technology foremost among them—and borrows heavily from aspects of Internet culture: forums, gaming, and social media, to name just a few. For example, the platform eBird, hosted by the Cornell Lab of Ornithology, functions like a Facebook for birders, allowing users around the globe to log their observations and compare their “life lists” of species sighted with those of others. Foldit, by contrast, has players compete in teams to win challenges and climb leaderboards.
There are as many varieties of citizen science as there are of science. In some fields, researchers look to citizen volunteers for help sifting through the deluge of information from microscopes, satellites, and telescopes. In other fields—like ornithology, where lay observations posted on eBird contribute to detailed maps of bird migrations—analytic capabilities have outstripped the available data, and scientists are asking citizens to gather more. Professionals may work side-by-side with small groups of dedicated amateurs in field experiments; alternatively, tens of thousands of citizen scientists participate from the comfort of their own homes, often in moments of boredom and procrastination.
“The common thread that runs through citizen science is that everyday people, who are not trained scientists, can contribute to science and be directly involved, that they understand basic research questions and want to help scientists answer those questions,” says Laura Germine, Ph.D. ’12, a postdoctoral researcher at Massachusetts General Hospital (MGH). She developed the website Test My Brain, which hosts psychological studies that have gathered more than 850,000 participants in the past five years (see page 57).
Yet for most scientists and laymen, that concept remains foreign. What, exactly, can untrained laypeople contribute to an endeavor as rarefied as scientific research?
Based on Baker’s work, the answer seems to be: a lot. In the five years since Foldit (fold.it) was launched, its more than 300,000 registered players (about 2,000 are active, playing more than once a week) can take credit for remarkable achievements. In one three-week challenge, they produced a near-exact model for a protein whose structure had eluded scientists for more than a decade. In another instance, they successfully redesigned an existing protein to increase its efficiency more than eighteenfold. Player strategies, in turn, have been studied by researchers seeking to improve computer algorithms, and Foldit now is challenging its users to design proteins that have never existed in nature. Foldit players—most of whom have little to no biochemistry background and who play the game in their spare time—are authors on four scientific papers, and their gameplay has contributed to several more.
The premise behind Foldit is that all human beings have advanced spatial-reasoning capabilities far beyond those of current computers, making protein-folding a visual and almost intuitive endeavor. As one top-ranked Foldit player told Nature in 2010, “It’s essentially a 3-D jigsaw puzzle.” “When you’ve got it right,” another player said, “you see your protein moving and changing shape, and your score rushes up. Your own player name rushes up through the ranks, and the adrenaline starts.”
In online challenges, an amino-acid sequence or partially folded protein is released to the entire Foldit community, and players work, usually in teams, to achieve the most stable configuration in the weeks or months allotted, swapping tips and frustrations in chat rooms and message boards. For the most part, Foldit seems like any other gaming community—apart from such objectives as “Hide the hydrophobics” and puzzles titled “Unsolved chicken anemia virus protein” and “Scorpion toxin.”
“Expert hydrogen bonding!” the program commends after a particularly successful move. “+396.”
“People are really smart,” notes Baker, who occasionally does Skype calls with players to answer questions or discuss improvements to the game. “The ones who get really into Foldit look at Wikipedia, and they learn a lot. The conversations you have with someone who has no scientific background at all, but has been playing Foldit for a while, are pretty high-level.” The lab’s Foldit support team regularly interacts with players through scientist chats and message boards. “I think it’s pretty critical to be responsive,” Baker says.
Citizen Computers
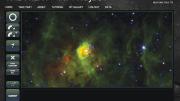
The 2007 launch of the citizen-science project Galaxy Zoo was met with immediate success: a site crash. Spurred by the enormous number of images captured by telescopes each day, astronomers from Johns Hopkins University and, in England, the University of Portsmouth and the University of Oxford had developed a website to involve amateurs in classifying galaxies based on shape—and the turnout stunned them. Initial traffic was 20 times what they had hoped for, and within 24 hours, online participants were tagging more than 60,000 images an hour. More than 150,000 people contributed more than 50 million classifications in the project’s first year.
“There are people who believe that computers are better than people at any task, if you’re just smart enough to program the computer properly,” says professor of astronomy Alyssa Goodman. “In truth, for nearly all pattern-recognition tasks, evolution has made the human brain very, very good—still better than any computer program.” Indeed, Galaxy Zoo represents a growing class of citizen-science projects that ask interested members of the public to do what computers still cannot. The citizen classifications, though useful, are not always ends in themselves. “There are tasks where, if you have a lot of people looking at data, then that trains the computer,” Goodman continues. “Then, the computer can do better than if you just tell it to find the solution.”
The new field of human computation aims to guide this integration of man and machine, combining inputs to tackle problems that neither humans nor computers can solve alone. Classically, computers have used entirely automated operations, but human computation involves tasks like image recognition or text analysis, where the exact process can be difficult to define through traditional programming commands. Rather than explicitly coding the characteristics of a galaxy, for instance, researchers are developing machine-learning methods that enable computers to infer the appropriate patterns from human-generated training sets.
“Astronomy is rapidly moving toward the regime where we’re going to have more data than we have any hope of manually looking at,” says Chris Beaumont, a software engineer at the Harvard-Smithsonian Center for Astrophysics. For his dissertation, he worked with Goodman to study interstellar “bubbles,” areas thought to be hotbeds of star formation. These bubbles, like galaxy shapes, are hard for computers to detect, but in an effort called the Milky Way Project, hosted by the citizen-science platform Zooniverse (an expansion of the original Galaxy Zoo effort; www.zooniverse.org), more than 35,000 citizen scientists identified more than 5,000 bubbles in images from the National Aeronautics and Space Administration’s Spitzer Space Telescope.
Beaumont has used these contributions to build more sophisticated algorithms for bubble identification that will cut down on the need for human input: for instance, a computer might screen large datasets and present lay volunteers and experts with only the most ambiguous cases. “If you’re looking for something that’s rarer, or if you’re looking through a much larger dataset, there aren’t enough people in the world to do what you need to do,” he says.
Moreover, after “learning” from so many amateur identifications, the algorithm can also distinguish between typical and suspicious lay contributions, providing a means to check users’ reliability and more accurately make use of data from citizen scientists. As Beaumont says, “We need to learn how to combine computers and humans to scale up to big data.”
Citizen Naturalists
Human computation frequently taps into a phenomenon called crowdsourcing: small contributions from a large base of users—in this case, citizens—can collectively accomplish huge tasks impossible for a small, dedicated group. At Harvard’s Center for Research on Computation and Society (CRCS), postdoctoral fellow Edith Law is developing an online citizen-science platform called Curio (www.crowdcurio.com) to crowdsource research tasks. (She plans to launch it this spring.)
She began by interviewing Harvard researchers across multiple disciplines. “I wanted to understand the opportunities,” she explains. “What bottlenecks do they have? How do they currently train people? Would they be comfortable sharing data, and at what stage? I was thinking about what crowdsourcing could bring to science.”
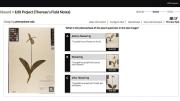
One faculty member she interviewed was Charles Davis, professor of organismic and evolutionary biology and co-director of the Harvard University Herbaria (HUH). He oversees one of Curio’s inaugural projects, which asks citizen scientists to help assess the ecological impact of climate change, and he is well aware of what amateurs can contribute. Together with Richard Primack ’72, professor of biology at Boston University, Davis found that spring flowering times in the eastern United States in 2010 and 2012, following unusually warm winters, were the earliest ever recorded—an average change of approximately three weeks within less than a century. The source of this historical comparison? Detailed records kept by naturalists Henry David Thoreau in Concord, Massachusetts, in the nineteenth century, and Aldo Leopold in Dane County, Wisconsin, in the twentieth—among the few sources of information on long-term ecological change. These valuable historical data gave the researchers detailed insight into the effects of climate change in the eastern United States over a 160-year time span.
But the work is far from done. “How do we gather the data we need to assess long-term climate change across all of New England?” Davis asks. Records like Thoreau’s and Leopold’s are rare, but he suggests another source of information—HUH collections, which contain nearly half a million samples from the region. In the new citizen-science project, Thoreau’s Field Notes, participants will be trained to classify digital images of herbaria specimens based on their phenophase (the visible stages in a plant’s life cycle, like budding or flowering). Davis hopes that linking these botanical markers with accompanying field notes—namely, the time and place of the specimen’s collection—will yield a more detailed understanding of climate’s effect on flowering time.
If the premise of the project—laypeople classifying images, whether plant specimens or interstellar bubbles—is beginning to sound familiar, Law would agree. Curio is built on the commonalities among disparate crowdsourcing projects. For instance, “You can think about these annotation tasks at a very abstract level,” she says. “Almost all annotation tasks have to do with describing objects or relationships between objects, either in an open-ended way” (describing an image using labels like “black” and “cat,” for instance) “or a close-ended way” (like classifying images into discrete, predetermined categories of “cats” or “dogs”).
But projects like Davis’s face the challenge of training citizen scientists to process complex information. “How do you identify a flower not just from a plant, but from a plant that’s flattened and on a piece of cardboard?” he asks. Many of his students, he says, are shocked when they encounter a specimen for the first time. “Presenting the untrained eye with these complex images and asking people to make sense of them is a real concern,” he continues, “and comes with its own set of challenges.” He and Law are designing a tutorial that will use labeled examples to train volunteers, and Curio is designed to integrate experts with a less-experienced crowd—for instance, controversial lay classifications may be sent to professionals for a final verdict.
Davis and Law hope the project will stimulate participants’ connections with the natural world. They plan to reach out to local gardening and naturalist communities for volunteers, and the aim is for amateurs to interface with both botanical specimens and timely research questions. “This work has certainly reached a broad audience locally,” Davis says. “It’s about organisms that people in this area know and love.”
Citizen Subjects
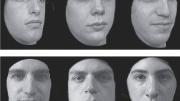
Scientific discoveries come from unusual places; widespread evidence of prosopagnosia, or face blindness, came from an online forum. Shared experiences draw people together all the time, but this common thread was something new: the inability to recognize faces. The phenomenon had been reported in the scientific literature, but almost entirely in connection with traumatic events like strokes. In the late 1990s, when groups of people online began describing entire lives spent recognizing acquaintances by their clothing or hair, they were at first dismissed—few researchers or clinicians had even heard of such a thing (see “Facial Pheenoms,” September-October 2009, page 7).
As the condition, known as “developmental prosopagnosia,” gained clinical and academic recognition, it swiftly captured the public imagination. While a research assistant at University College London from 2005 to 2007, Laura Germine of MGH helped develop a test of face recognition that ran online, not in the lab, to accommodate rapidly increasing public interest. Before long, tens of thousands of people were participating. Most did not think themselves face-blind; they were simply curious about how they measured up.
“People want to do these things,” says Germine. “Learning about yourself—learning about your personality, learning about what you’re good at, learning about what you’re less good at—is something people are very interested in doing.” Inspired by the strong reception the facial-recognition tests received, she developed Test My Brain (testmybrain.org) in 2008 as a platform to host psychological studies. Participants take short tests with names like “Famous Faces” and “Holding Information in Mind” in return for personal feedback and a description of the scientific research involved.
Yet many researchers were initially skeptical about data—especially of the sort requiring precisely timed responses—gathered in the unsupervised setting of the Internet. Unless scientists used recruited and compensated volunteers who were tested under carefully controlled conditions, how was it possible to know that subjects were not cheating, lying, or simply becoming distracted? In response, Germine and colleagues published a study in 2012 that compared data from Test My Brain with data from studies conducted using traditional methods. Though the much larger Web samples showed slightly higher variance, the researchers found no consistent differences in other aspects of performance or data quality.
Web data, in fact, may have unique advantages, thanks to the diversity of its participants. “Most research in the world happens on campuses in the United States, so what we know a lot about is undergraduates in the United States,” says Josh Hartshorne, Ph.D. ’12. “They’re diverse in some ways and homogenous in others.” Hartshorne, now a postdoctoral fellow at MIT, runs a website called Games With Words (gameswithwords.org) that hosts language experiments. Recently, he says, researchers have become aware of the possible pitfalls of generalizing results derived from participants that some psychologists now dub “WEIRD”—Western, educated, industrialized, rich, and democratic. The enormous sample sizes of Web data, on the other hand, can in fact help characterize cultural differences in areas like cognition and social behavior; for example, researchers from the CRCS have used an online platform called Lab in the Wild to quantify cultural preferences for website aesthetics.
“We have these new technologies,” Hartshorne explains. “What can we do with them that we couldn’t do before? That’s what we should be doing. There’s this unexplored territory where we can make very rapid progress.” For instance, Germine explains, Web data are galvanizing the field of differential psychology—the study of individual differences rather than common basic mechanisms—and ordinary citizens, with the help of online tests, are increasingly able to characterize themselves for their own and for researchers’ benefit.
“I think there’s a shift now, in medicine and every other domain, toward wanting to learn about yourself and having that be in your own hands,” says Germine. “Increasingly, knowledge is available on the Internet, and people can interpret that themselves” (as with developmental prosopagnosia). “There’s a much higher ability to take things into your own hands, for better or for worse.”
Citizen Patients
Some members of the medical community are beginning to take note. Patients with chronic illnesses, for example, are frequently forced to become experts on their own conditions. “In a week,” says Eva Guinan, associate professor of radiation oncology at Harvard Medical School (HMS) and associate in medicine at Boston Children’s Hospital, “patients could put together a profile of what living with a disease is like that I could never attain as a practitioner.”
Advances in DNA sequencing technology have made genetic information plentiful, but data about symptoms and disease outcomes remain in relatively short supply. Here, the public can help, says Stephen Friend, a former HMS faculty member. He believes that citizens, in addition to going into forests or backyards to collect data, can help research by gathering information on themselves.
As president of the nonprofit Sage Bionetworks, based at Seattle’s Fred Hutchinson Cancer Research Center, Friend is developing a platform to engage patients in collecting and interpreting their own medical data. One of his newest projects, undertaken in collaboration with Guinan and the Fanconi Anemia Research Fund (a patient-support and fundraising group), focuses on a rare, genetic blood disorder that puts patients at high risk of head and neck tumors. “If you ask [Fanconi anemia] patients what they’re worried about,” he explains, “they’ll say, ‘Can you tell me what puts me at risk? Can you tell me ways to find it early?’”
These tumors’ causes are still poorly understood; though there is a genetic component, the environment likely plays a role as well. Instead of having patients see a doctor once or twice a year, Friend continues, “we’re getting them trained to take photographs of their own mouths,” where cancers frequently appear, “and to give narratives of what they’re doing”—stress or eating patterns, for instance. Patient self-monitoring, in addition to helping catch tumors early, may also contribute to medical research: Friend suspects that these patient journals may hold clues to understanding the course of the disease. Following an “open science” model, Sage Bionetworks will make the data publicly available online and challenge researchers worldwide to “turn anecdotes into signal.” (See the Web Extra, “More Shots on Goal,” to learn more about crowdsourced innovation.)
Another project aims to use the popular iSleeping mobile phone app to gather data on the effect of sleep medications. The app, developed by researchers in France, already monitors the sleep patterns of more than 600,000 people by analyzing snoring and user movement, effectively creating an automatic sleep log. Friend hopes to enroll 1,000 users in a clinical trial that will make use of this detailed data. Other initiatives are taking similar approaches to soliciting patient contributions. The Personal Genome Project, headed by Winthrop professor of genetics George Church at HMS, asks people to make their genome sequences available for medical research. The American Gut Project sends participants a kit with which to sample the bacteria living on and in their bodies; a related effort has recruited more than 1,000 volunteers to test the microorganisms in their homes.
Friend believes these efforts increase citizens’ and patients’ stake in biomedical research that otherwise can feel distant. “You have citizens who are willing to do extraordinary things to treat themselves,” he observes. The question now for the research and medical community, Friend says, is: “How do you get the public nurtured as full partners?”
Citizen Ownership
Yet the road to full partnership brings additional challenges. Fields like human computation are exploring how best to utilize lay participation and integrate it with traditional research, but citizen science in the Internet age carries all the ambiguities of the digital world—concerns about trustworthiness, privacy, intellectual property, the role of expertise in the age of Wikipedia. As citizens assume more involved roles, these issues grow progressively more complex. Could patients withdraw personal information they’ve collected and donated? Who would own a protein that a team of Foldit players helped design?
One major question facing citizen science is that of citizen ownership. Leaving aside questions of authorship and intellectual property, amateur contributions to science tend to be narrowly circumscribed. “Lay participation presupposes that somebody else knows the scientific value of the thing being studied,” says Sheila Jasanoff. “Even when the lay citizen is listening to birdsong or going on expeditions into the woods each spring to catch a glimpse of what migratory birds are around, that citizen is not determining the population-movement charts that the ornithological community is creating out of those observations.” Though initiatives like Foldit and Zooniverse have resulted in multiple scientific publications—with some citizen scientists as coauthors, in Foldit’s case—the intellectual work of analysis and interpretation still rests, ultimately, with trained professionals.
Some citizens find unusual ways to make projects their own: Germine and Hartshorne, for instance, report that classroom teachers sometimes ask students to interpret the personal feedback scores from Test My Brain and Games With Words, or collect the scores as data sets for classroom analysis. The researchers themselves receive feedback: participants often critique the study design or suggest their own interpretations of results. “Every participant is like a mini-reviewer,” says Germine. “Ordinary people can provide a lot of insight into your own data that you and your colleagues would never have thought of.” Likewise, CRCS’s Edith Law suggests that citizen science can educate amateurs about the realities of scientific research, warts and all, by exposing them to data and data processing. “It can teach people what scientists do,” she says, “and how they analyze problems.”
Other projects push the bounds of citizen participation. Public Lab, an initiative of the MIT Center for Civic Media, takes a do-it-yourself approach to involving citizens in environmental science. An amateur “biohacker” movement applies a similar ethos to inexpensive, self-guided genetic engineering, and it has occasionally clashed with police and the Federal Bureau of Investigation over issues of safety. The Internet has begun democratizing science in surprising ways; some researchers make comparisons to how personal computers have altered technology and society. Web-based research, says Hartshorne, “is maybe the equivalent of a kid in his or her garage, inventing the next big tech company.”
But for the most part, the question remains: is citizen science intended ultimately for the citizens or the scientists? The very reason for the growing popularity of citizen science—its usefulness in research endeavors—may paradoxically diminish the quality of engagement for its lay participants. Bluntly put, in a time of tight federal funding, lay participation is cheap. According to a 2012 study from the University of Maryland, “scientists saw citizen-science projects mainly as an opportunity to facilitate large-scale data collection,” though “altruistic” motivations like increasing scientific literacy were also named. Law points out that in most online projects, the scientists have never met their citizen participants. “What would happen if we had a conference of citizen scientists?” she asks. Technology may have provided citizen science with diverse avenues to narrow the gap between amateurs and experts, but further progress—if that is indeed the movement’s goal—will require dedicated effort on both sides.
Most researchers involved with citizen science believe this vision is one worth seeking, whatever the way forward may be. “To what degree does citizen science bring the lay community closer to the interface of science and society?” asks Eva Guinan. “In a world where so many people say and feel that they are being left behind by science and technology, does citizen science help? Or does it act like just another online game?”