S
ynthetic biology, or the application of engineering principles to the design of life, presents world-changing prospects. Could components of a living cell function as tiny switches or circuits? How would that allow biomedical engineers to build biological “smart devices”—from sensors deployed inside the body to portable medical kits able to produce vaccines and antibiotics on demand? Could bacterial “factories” replace the fossil-fueled industries that produce plastics, foods, and fertilizers? Will the secrets of living creatures that enter suspended animation during periods of drought and extreme cold be harnessed to keep human victims of trauma alive? And is the genetic information preserved in long-frozen or fossilized extinct species, like woolly mammoths, sufficiently recoverable to help save living species?
These ideas, once the stuff of science fiction, are now the stuff of science. Some aren’t yet functioning realities, but others have launched business applications, whether in medicine (such as hospital gowns that signal exposure to infection) or in land remediation (where bacterial “factories” powered by the sun capture nitrogen from the atmosphere to help plants grow). Someday, engineered forms of life that store carbon may even be one of the solutions to Earth’s climate-change problem.
“Most of biology, historically, has been analyzing how nature works,” says Donald Ingber, director of Harvard’s Wyss Institute for Biologically Inspired Engineering. Systems biology is the culmination of that effort to deconstruct natural processes. Now, with synthetic biology, he points out, scientists “are at the point where we know enough that we can actually engineer artificial and natural biological systems.” Researchers today can build things from biological parts, and even create hybrid systems by linking them to non-living machines.
Propelling the science forward are scores of innovations in biological science, with new discoveries coming every month. Among the most important are advances in genetic editing, including improvements in accuracy, and the ability to make hundreds of changes at once. Another is computer-aided design, widely used to model biological systems, and to build new proteins by combining amino acids in ways never seen in nature. Advances in molecular engineering are driving the construction of ever-more complex circuits made from biological materials. And the ability to freeze-dry cellular transcription machinery outside the confines of cells has enabled scientists to easily manufacture proteins at will, at any time and place.
Most synthetic biology today centers on single-celled organisms, such as the bacterium Escherichia coli. In the spring of 2019, for example, researchers at the University of Cambridge announced that they had synthesized an entire E. coli genome—the bacterium’s complete set of genetic material—and swapped it into living E. coli cells. Their version, which at four million base pairs (the building blocks of the DNA double helix) was then the largest synthetic genome created to date, was nevertheless a pared-down variant that eliminated redundancies, thereby simplifying the biological complexity evolved over millennia and making the organism easier for human engineers to understand and manipulate. Although the bacteria with synthetic genomes were not as robust as their natural counterparts, and reproduced slowly, they survived.
The scientists who do this work know that they are operating in a new domain, with transformative possibilities. James Collins, a core faculty member at the Wyss Institute who 20 years ago created one of the first biological circuits, believes this synthetic, engineered biology “will be a defining, if not the defining, technology of the century.”
Snapshots of the evolving technologies follow.
Switches, Sensors, and Medicines on Demand
In January 2000, Collins and a team at Princeton, working separately, simultaneously published in Nature the first designs for switches made from biological parts. That innovation arguably marked the beginning of modern synthetic biology. Collins had engineered a switch, like the push button on a desk lamp: press once, the light turns on; press again, it turns off. Collins’s simple mechanism was not built from metal and plastic, but from two genes that flip-flopped between on and off states when stimulated by chemical signals or changes in temperature. It was a simple circuit.
The simplicity of what Collins had created belied its potential. The ability to engineer biological circuits in this way meant that cells could represent binary states such as the zeros and ones that are the basis of computer systems. They could perform simple logic. And because they could be programmed to die after a certain number of cell divisions, they enabled the creation of the first kill-switch safety mechanisms to prevent organisms with synthetic parts from escaping into the environment.
Many of the earliest biological switches were crude and prone to accidental triggering. The inside of even a single-celled bacterium such as E. coli, where engineered synthetic circuits are often introduced and tested, is very busy. There are “many molecules, large and small, interacting in a very small space,” explains Collins, who teaches in the Harvard-MIT Program in Health Sciences and Technology. All this activity can lead to what engineers call “crosstalk,” instances when a stray signal can accidentally flip a simple switch. Such unpredictable behavior would be anathema in biomedical applications.
But biological switches have become much more robust since 2000, allowing them to be used in laboratory animals as reliable detectors that signal the presence of pathogens. In applications in the gut, for example, RNA-based switches have been designed to release appropriate probiotic therapies. Such an RNA-based switch takes advantage of the fact that the bases on a single strand of RNA want to pair with opposite bases, just as “A[denine] goes to T[hymine] and G[uanine] goes to C[ytosine]” in DNA base pairing, Collins explains. This means that an RNA probe can be designed to combine systematically with an opposite RNA strand associated with a bacterial pathogen. That opens the switch, which then releases a fluorescent particle that reveals the presence of the pathogen.
To effect this, researchers must engineer biological circuits and insert them into living bacteria: a process that sounds formidably complex. But scientists know that different kinds of bacteria, in the course of evolutionary history, have routinely exchanged whole “cassettes” of many genes, such as those that control metabolism. This swapping of genetic material is called “horizontal gene transfer.” Within these cassettes that control particular biological functions, researchers have devised ways to alter specific genes, and then reinject the whole functioning circuit into a cell.
These genetic circuits control what proteins the cell produces. Since the 1960s, biologists have known that the protein-synthesizing machinery of a bacterium can even be plucked from within the cell’s protective outer membrane, placed in a laboratory test tube or petri dish, and still function.
The lab’s first deployment of the technology was to create a detector for Ebola, a rare but deadly disease that has killed thousands of people in West Africa.
In October 2014, the field took another leap forward when the Collins lab published a serendipitous discovery that advanced the practicality of using RNA-based switches as detectors in the field—and even as therapy-producing agents. While working to develop encapsulated cell-free genetic networks for cellular reprogramming, postdoctoral student Keith Pardee discovered that a cell’s transcription and translation machinery (the parts that build proteins from DNA instructions) could be spotted onto a piece of paper, freeze-dried, and then restored to full function when rehydrated. The lab’s first deployment of the technology was to create a detector for Ebola, a rare but deadly disease that has killed thousands of people in West Africa. Subsequent testing revealed that their Ebola detector remained viable for a year or more—without the need for refrigeration. This was significant, because Ebola often strikes in remote regions poorly served by healthcare facilities.
The lab has subsequently used the same techniques to make diagnostics for the Zika virus, for gut microbiome analysis, and for detecting antibiotic resistance. “We now have efforts underway looking at Lyme disease, HIV, TB, HPV, and hepatitis C,” says Collins. “It has provided us with a whole new diagnostic platform.”
Because these paper-based tests contain no living cells, he points out, there are neither biocontainment safety concerns nor problems of storage and distribution. “You have marvelous sensors” made from “biological components that are incredibly inexpensive, easy to use, and rapid”—providing results in less than an hour.
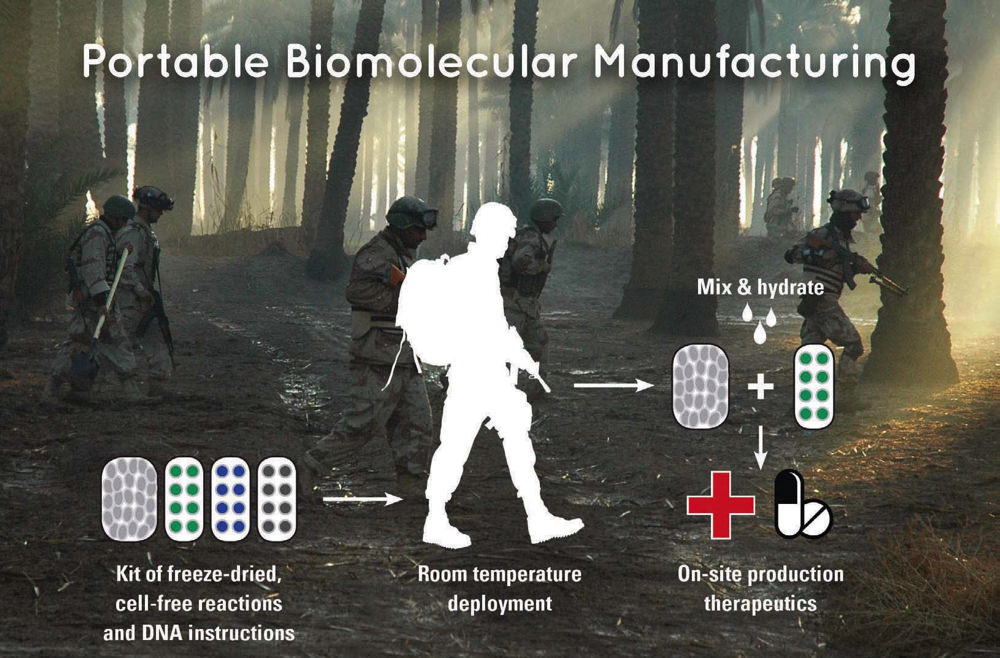
The ability to freeze-dry the mechanisms that cells use to produce proteins has led to portable diagnostics and therapies, such as vaccines and antibiotics that can be manufactured on demand in the field.
Image courtesy of James Collins/MIT/AB Forces News Collection/Alamy Stock Photo
Collins’s lab went on to demonstrate that this transcription-translation machinery could be spotted and freeze-dried into fibers used to make clothing, and is now completing work on wearable synthetic biology: suits that could show first responders or military personnel whether they have been exposed to a nerve toxin; johnnies that could tell whether a patient has a bedsore or an antibiotic-resistant infection; and lab coats that could signal whether a physician has been exposed to dangerous MRSA (methicillin-resistant Staphylococcus aureus, an antibiotic-resistant form of staph) or Clostridium difficile pathogens. (The lab recently won a Johnson & Johnson contest to design the lab coat of the future.)
Collins has also expanded the uses of freeze-drying to portable therapeutics. Just as the cellular machinery of a bacterium can be made to detect a pathogen, it can be engineered to produce a vaccine or an antibiotic such as vancomycin.
Most antibiotics are products of soil-dwelling bacteria. To make a therapeutic such as vancomycin, Collins takes existing circuits (or bits of DNA) that “are found naturally or have already been produced by others,” that will “make the molecule of interest.” He uses gene-editing tools such as CRISPR to insert these circuits into cells, and then freeze-dries these circuits together with the machinery for transcription and translation—the parts that enable that modified DNA with its associated genes to be “turned on and express the relevant proteins.” In the case of diagnostics, the rehydration that activates protein production might be a patient’s blood, urine, or sputum. In the case of therapeutics, adding water is enough to revive the machinery for making vaccines or antibiotics. This has enabled his lab to develop field kits the size of a small cellphone that could be carried “by soldiers or hikers or astronauts, athletes, or healthcare workers in global healthcare settings” to make their own medicines when needed. The individual would take the freeze-dried contents of a cell (its protein-manufacturing apparatus), add water to restore the bacterial machinery’s ability to synthesize proteins, and then add the freeze-dried engineered circuit containing the DNA instructions for making “a vaccine or an…antibiotic”—and within a couple of hours at body temperature, the therapy “is there to be used in the field.”
Taking advantage of the diversity and power of biology in this way, Collins says, “enables you to move synthetic biology out of the lab.”
Bacterial Factories of the Future
Pamela Silver, a founder of the field of synthetic biology, runs one of Harvard’s most prolific labs, with dozens of projects in train at any time. Among the most important questions she faces, therefore, is what to do next. For a long time, says the Adams professor of biochemistry and systems biology, synthetic biology was producing “toy systems. But now we’re at a point where we’re saying, ‘Forget this. We actually need to solve real-world problems.’”
Agriculture is one area of critical need: a growing world population must be fed. Silver and her long-term collaborator Daniel Nocera, Rockwood professor of energy in the department of chemistry and chemical biology, have therefore devised a process to manufacture inexpensively one of the most energy-intensive products farmers use: fertilizer, which provides the nitrogen that plants need to grow.
Pamela Silver
Photograph by Jim Harrison
The work builds on an artificial leaf invented earlier by Nocera, a device that harnesses solar energy to split water (H2O) in order to produce hydrogen, an energy source. To create fertilizer, Silver helped Nocera and members of his lab connect the artificial leaf to a strain of bacteria that can “fix” both atmospheric carbon dioxide and nitrogen, converting them to organic forms that can be used by living organisms. Provided with an unlimited supply of hydrogen from the artificial leaf, the bacteria combine the hydrogen with carbon pulled from the atmosphere to create a solid fuel that the bacteria store internally, as a long-term energy supply. “You can imagine this as making the bacteria fat,” explains Nocera: “obese, solar-fed bacteria” that are up to 30 percent stored energy by weight.
The bacteria are then mixed into the soil, where some remain, and others form associations directly with plant roots. Drawing from their stored energy reserves, they begin fixing atmospheric nitrogen, thus fertilizing the plants. (Because the carbon in the bacteria remains sequestered in the soil even after the bacteria deplete their energy stores and die, the process has the added advantage of being atmospheric carbon-negative.) On test fields, Nocera reports “big increases in crop yields” with almost no run-off, an environmentally poisonous side effect of water-soluble chemical fertilizers.
Turning bacteria into plant food that can be applied to fields “has huge implications,” Silver explains, “because you can produce the bacteria locally and do so in a completely ‘green’ production cycle.” The system has been spun off to a company, Kula Bio, that produces low-cost, organic fertilizers.
Silver’s latest, “craziest,” venture, as she puts it, is a project to study suspended animation—biostasis—as part of a DARPA-funded (Defense Advanced Research Projects) research program. Her team, one of four, received more than $14 million to develop ways to slow metabolic processes in order to lengthen the time a trauma victim, whether soldier or civilian, can survive without medical help.
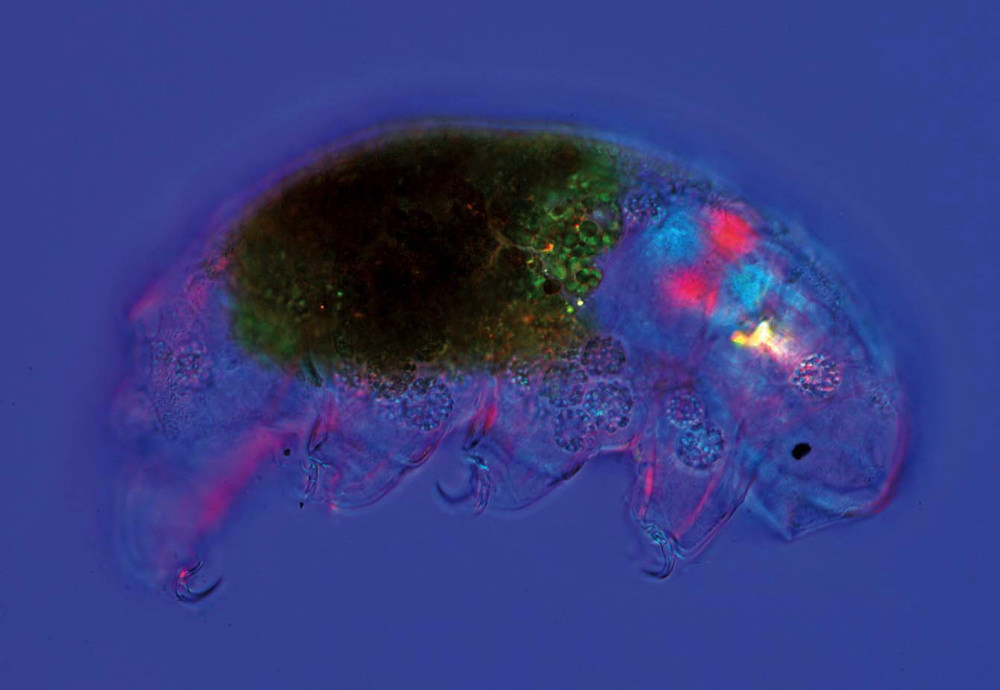
Tardigrades, or water bears, can survive extremes of drought, heat, and cold for as long as a decade in a state of metabolic suspension.
Image by Marek Mis/Science Source
To do so, she is studying tardigrades: water-loving creatures roughly two-thousandths of an inch long that live everywhere from oceans to mountaintops. Sometimes called moss piglets, these organisms can withstand extreme temperatures, drought, and even cosmic radiation, achieving extraordinary feats of survival by entering a state in which they shut down metabolism almost entirely. This trick, known as cryptobiosis, conjures images from science fiction of suspended animation during human flights to distant colonies on other planets.
Extremophiles like the tardigrade, Silver explains, commonly carry what early researchers called “intrinsically disordered proteins,” possessing a structure that appears to be mutable and gel-like. When facing desiccation, tardigrades use these proteins, mixed with sugars, to form a protective glassy film that fills their cells and encapsulates their organs. Silver seeks to engineer a synthetic version well suited for use with human cells. To achieve this, she has teamed up with associate professor of systems biology Debora Marks, a computational biologist and co-principal investigator on the project, who will use a machine-learning algorithm to suggest candidate protein designs that might work in human tissues.
“How do you make a world for 10 billion people? The only way you are going to do that is by engineering biology.”
Intrinsically disordered proteins could eventually help reduce the severity of heart attacks by limiting heart-muscle damage, or the impact of a stroke by slowing the death of nearby neurons. They might also extend the viability of human eggs stored for use in fertility treatments, or lengthen the usable life of organs harvested for transplant. For now, though, Silver acknowledges, “It’s early days.”
And when asked about the biggest challenge for synthetic biology, Silver replies bluntly that the true challenge is “How do you make a world for 10 billion people? The only way you are going to do that is by engineering biology”—work that is just beginning.
Applying Ancient DNA to Contemporary Conservation
Could synthetic biology save one elephant species by reanimating at least a portion of the biodiversity of an extinct species? George Church, perhaps the pre-eminent genome technologist in the world, believes so.
Church does basic-science research as well as fundamental technology development, working at the nexus of Internet technologies, computing, and biotechnology. Broadly speaking, he says, synthetic biology is molecular engineering: whether it utilizes biology to simulate logic circuits, probe the origins of life (a topic covered in depth in “How Life Began,” July-August 2019, page 40), manipulate metabolism, manufacture products, or enhance biological imaging. As an engineering discipline, says the Winthrop professor of genetics, “biology affords you the use of very sophisticated parts. Way more sophisticated than the piston in a car or the transistor in a phone. Biological parts have been through billions of years of debugging—trial and error.”

George Church
Photograph by Jim Harrison
In synthetic biology, this trial and error process is even better than the real thing because it can be accelerated, he explains. “We can make a trillion prototypes and then design a system that will select the best” (see “Harnessing Evolution,” January-February 2017, page 15, for a description). Church’s lab runs on a similarly accelerated pace that frequently puts his forward-looking projects in the public domain.
News accounts often report that he wants to revive the woolly mammoth, a cold-adapted elephant that roamed Siberia as recently as 10,000 years ago. In fact, Church is undertaking a multi-pronged conservation project—with synthetic biology at its core—that he hopes will benefit living Asian elephants, humans, the Arctic, and perhaps the planet.
Asian elephants carry endotheliotropic herpesvirus (EEHV), which kills up to 80 percent of newborns that contract it. The virus has caused hemorrhagic bleeding and death in numerous Asian elephants held in captivity, and recent testing in Southeast Asia points to it as the cause of documented wild-elephant deaths there as well. To help save this endangered species, Church plans first to edit its genome to make it resistant to EEHV or to all viruses.
Doing so will require multiple edits in several parts of the elephant genome. But that part won’t be difficult, he says: Church’s team (Eriona Hysoli, Bobby Dhadwar, and Jessica Weber) has perfected the ability to engineer virus-free mammalian genomes as part of a virology program for breeding pigs whose organs can be safely transplanted into humans. The hard part is the real-world problem of how to respond to the elephants’ shrinking habitat. “It’s not just the herpesvirus that’s putting them at risk,” he explains, “it’s their proximity to human beings. An elephant will innocently stamp on farmers’ gardens and destroy a year’s crops, and so the farmers, correctly according to their own ethics, say, ‘Well, that elephant owes us some food.’ And they kill the elephant that night.”
He has a genetic solution to that problem, too. He proposes further edits to the elephant’s genome that would introduce woolly mammoth genes into Asian elephants in order to confer cold resistance.

Woolly mammoth adaptations to frigid environments included blood oxygen release at low temperatures, the growth of thick hair, and the accumulation of subcutaneous fat for insulation and fasting.
Illustrations by Ben Novak/Courtesy of Revive & Restore
Church’s team began the process by comparing the genomes of Asian elephants to those harvested from the remains of woolly mammoths, which are about 99 percent similar, he says. By focusing on the differences, he can home in on those that enabled woolly mammoths to survive in places like the Siberian tundra. Like all genes, those that allow organisms to survive in such inhospitable environments are largely conserved across species, making it possible to determine the function of most genes without having to start from scratch. As for the few genes that are completely new, Church has numerous genetic tools at his disposal to determine their function. He can splice genes into other organisms to observe their effects, or silence them to see what changes then take place, or induce mutations in vitro, to see how that changes the protein products of the altered genes.
He has already introduced genes recovered from mammoth DNA as much as 700,000 years old into cultured cell lines. Some facilitate blood oxygen release at low temperatures, others the growth of thick hair and the accumulation of subcutaneous fat. Endowed with these genetic gifts, the potential range of the Asian elephant would expand into frigid areas where the animals would be far less likely to compete with humans for habitat. “We’ll be further increasing their diversity, possibly even making them more genetically diverse than any species has been before in that lineage,” Church explains, “because we’re not limited by time or geography, or even by natural DNA.”
Church has successfully demonstrated the function of two woolly mammoth gene variants that aid adaptation to a cold environment and has plans to modify at least 44 more elephant genes.
The idea is not so farfetched as it might sound: “There’s evidence that there was interbreeding in the past,” he says. “It’s like the evidence in the human genome that our ancestors interbred with Neanderthals—it’s a pattern with elephants and mammoths.” Besides recently developing (with professor of genetics Chao-ting Wu) an improved method for reading ancient DNA, Church has successfully demonstrated the function of two woolly mammoth gene variants that aid adaptation to a cold environment, and has plans to modify at least 44 more elephant genes based on what he has learned from mammoths and other sources.
His approach is also pragmatic: he has thought through business models that might support this effort. “Bison,” he points out, “returned from near extinction to a population of almost half a million worldwide, largely because the species has very low-cholesterol meat. Mammoths, or cold-resistant elephants, could support even more business models: you’ve got tourism, meat, hair (following a sheep model of seasonal removal), and maybe legal ivory. So there’s a lot more models for cold-resistant elephants than for bison.”
Church is sanguine about the potential of biological engineering to make a better world. “It’s easy to get overexcited…because everything that’s currently made by non-biological methods—inorganic materials—could be made by biological systems: bacteria make magnets, sponges make fiber optics, they make hard shells, bones, all sorts of inorganic materials.” Such optimism helps explain why synthetic biologists dot the departments of genetics and systems biology at the schools of medicine and engineering and applied sciences. As a transdisciplinary field, synthetic biology almost demands collaboration; more than a few labs do so through the Wyss Institute, and Church’s lab does, too.
The near-term future of synthetic biology, and particularly commercial applications, are being shaped as much by market forces (including social acceptance) as by technological advances within the field’s core disciplines of molecular and genetic engineering, whole-cell, cell-extract, or organism-level tinkering, and computer modeling. Although the ethics of human enhancement and environmental interventions will be debated, the often life-saving applications of synthetic biology to medicine and health are not only profitable but embraced by the public, presumably ensuring their steady growth. (Sales of the 10 biggest “blockbuster biologics”—commercial drugs with more than a billion dollars in sales that are manufactured with the use of living organisms—already exceed $70 billion annually in the United States, and are obvious candidates for improvement.) And immunotherapies—modifications to immune cells that enable them to recognize and fight diseases such as cancer—are an area of intense research.
“I think there’s almost no limit to synthetic biology,” Church says. “It will help us build space colonies, it will help us build anything that’s atomically precise. Everything, including circuits, would be better if it were atomically precise—and we can make things that way. Because that’s what biology does very well.”